Test for linear or nonlinear collinearity/correlation in data
collinear(x, p = 0.85, nonlinear = FALSE, p.value = 0.001)
Arguments
| x | A data.frame or matrix containing continuous data |
|---|---|
| p | The correlation cutoff (default is 0.85) |
| nonlinear | A boolean flag for calculating nonlinear correlations (FALSE/TRUE) |
| p.value | If nonlinear is TRUE, the p value to accept as the significance of the correlation |
Value
Messages and a vector of correlated variables
Details
Evaluation of the pairwise linear correlated variables to remove is accomplished through calculating the mean correlations of each variable and selecting the variable with higher mean. If nonlinear = TRUE, pairwise nonlinear correlations are evaluated by fitting y as a semi-parametrically estimated function of x using a generalized additive model and testing whether or not that functional estimate is constant, which would indicate no relationship between y and x thus, avoiding potentially arbitrary decisions regarding the order in a polynomial regression.
Author
Jeffrey S. Evans <jeffrey_evans<at>tnc.org>
Examples
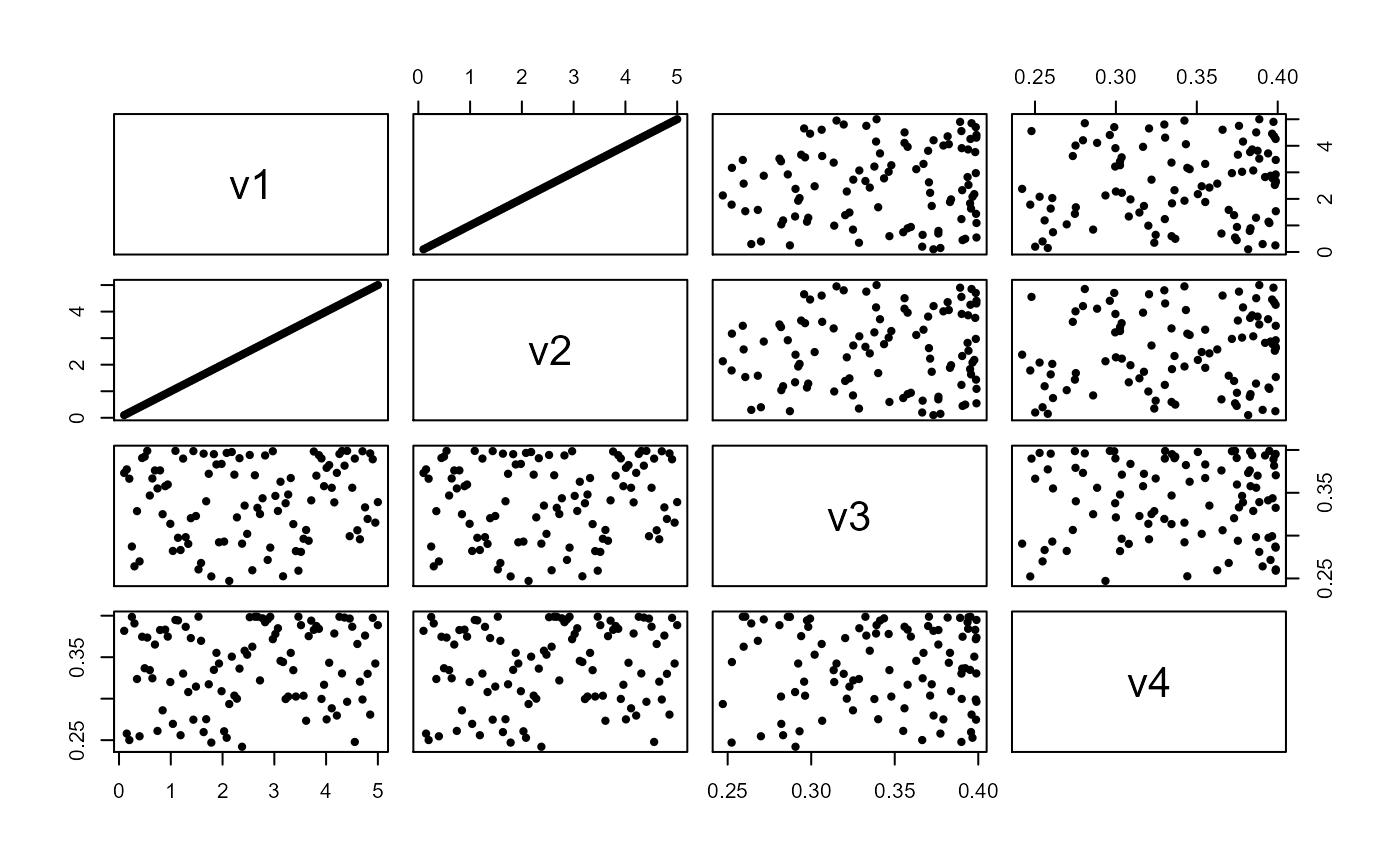
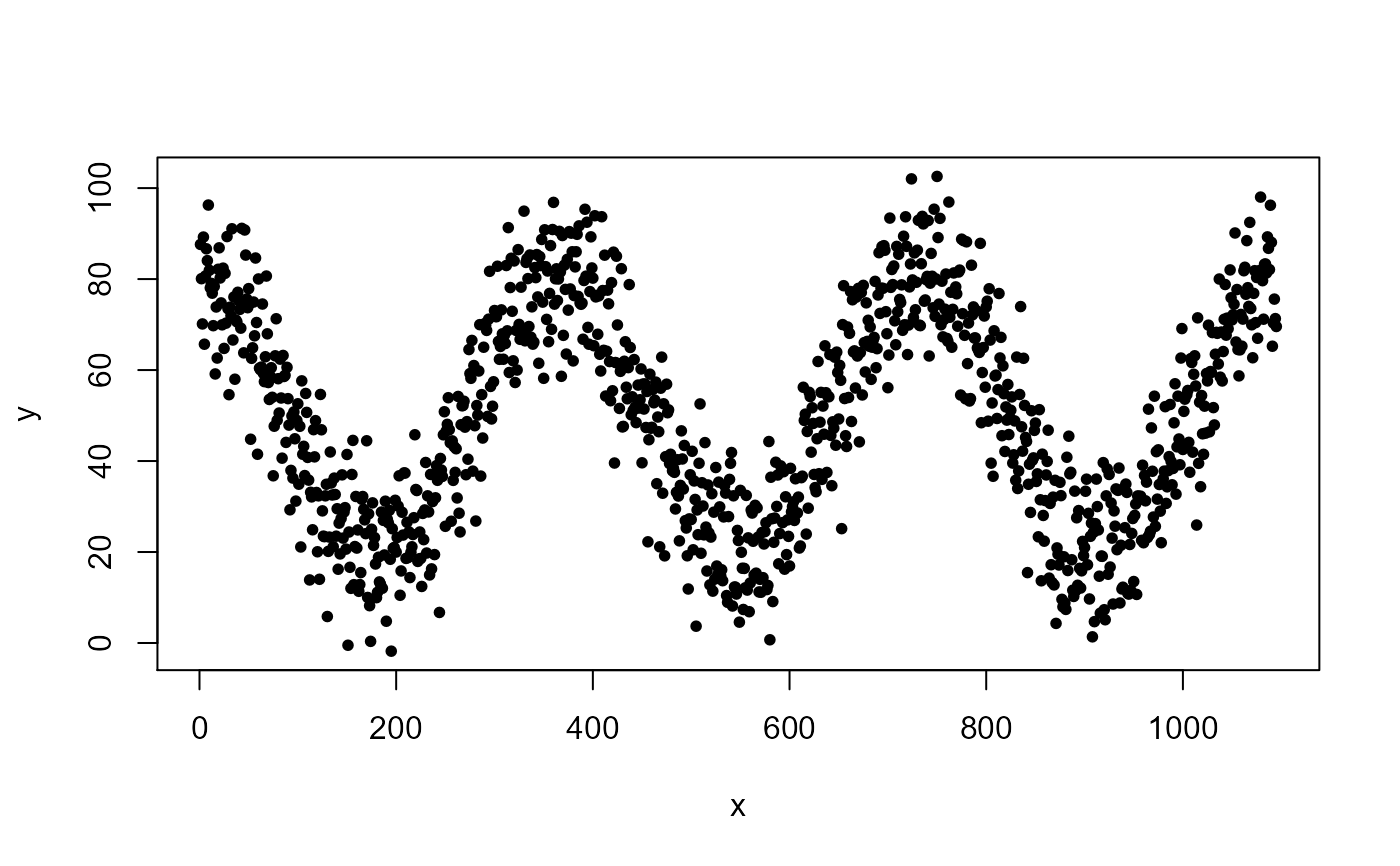
data(cor.data) # Evaluate linear correlations on linear dataCollinearity between head( dat <- cor.data[[4]] )#> v1 v2 v3 v4 #> 1 0.1000000 0.1000000 0.3731468 0.3817370 #> 2 0.1494949 0.1494949 0.3773337 0.2579079 #> 3 0.1989899 0.1989899 0.3664366 0.2501137 #> 4 0.2484848 0.2484848 0.2872536 0.3985506 #> 5 0.2979798 0.2979798 0.2641251 0.3906866 #> 6 0.3474747 0.3474747 0.3285911 0.3237098( cor.vars <- collinear( dat ) )#>#>#> #>#> [1] "v1"#> v2 v3 v4 #> 1 0.1000000 0.3731468 0.3817370 #> 2 0.1494949 0.3773337 0.2579079 #> 3 0.1989899 0.3664366 0.2501137 #> 4 0.2484848 0.2872536 0.3985506 #> 5 0.2979798 0.2641251 0.3906866 #> 6 0.3474747 0.3285911 0.3237098# Evaluate linear correlations on nonlinear data # using nonlinear correlation function plot(cor.data[[1]], pch=20)collinear(cor.data[[1]], p=0.80, nonlinear = TRUE )#>#>#> #>#>#> [1] "x"